Stage M2 : AI-powered multimodal integration of spatial omics data
Glioblastoma (GB) is an aggressive brain cancer, characterized by extensive infiltration and metabolic adaptations, yet not fully understood. Spatial transcriptomics (ST) and spatial metabolomics (and lipidomics) through Mass Spectrometry Imaging (MSI), are two powerful omics modalities that allow to dissect the tissue organisation of this cancer. Our bioinformatics team, led by Dr Nikolski, in collaboration with Dr Daubon’s cancer biology team, studies GB metabolism using ST and MSI data, developing dedicated and innovative computational approaches. We (Nikolski’s lab) have already developed SpacePath (pathway activity under ion–metabolite probabilistic framework for MSI; manuscript in submission) and DIMet [1, 2]. These tools provide an immediately usable software and modelling base. We have in-house paired ST + MSI datasets on consecutive PDX sections, and also public paired ST/MSI datasets [3]. Now we move towards multi-modal integration.
Despite recent advances in multi-modal spatial omics integration, metabolic semantics are still poorly used by existing frameworks: the key molecular modality (MSI-based spatial metabolomics/lipidomics) differs from ST in resolution, quantitativeness and section-to-section registration [4]. Most ST–MSI integration across adjacent sections relies on geometric registration (histology-derived anchors), however, invasive cells are sparse and white-matter anatomy is anisotropic, which can lead to biological signal misalignment. On the other hand, graph-based fusion methods (e.g., SpatialGlue, SpatialMET, COSMOS) improve integration in more homogeneous settings.
Work for the internship
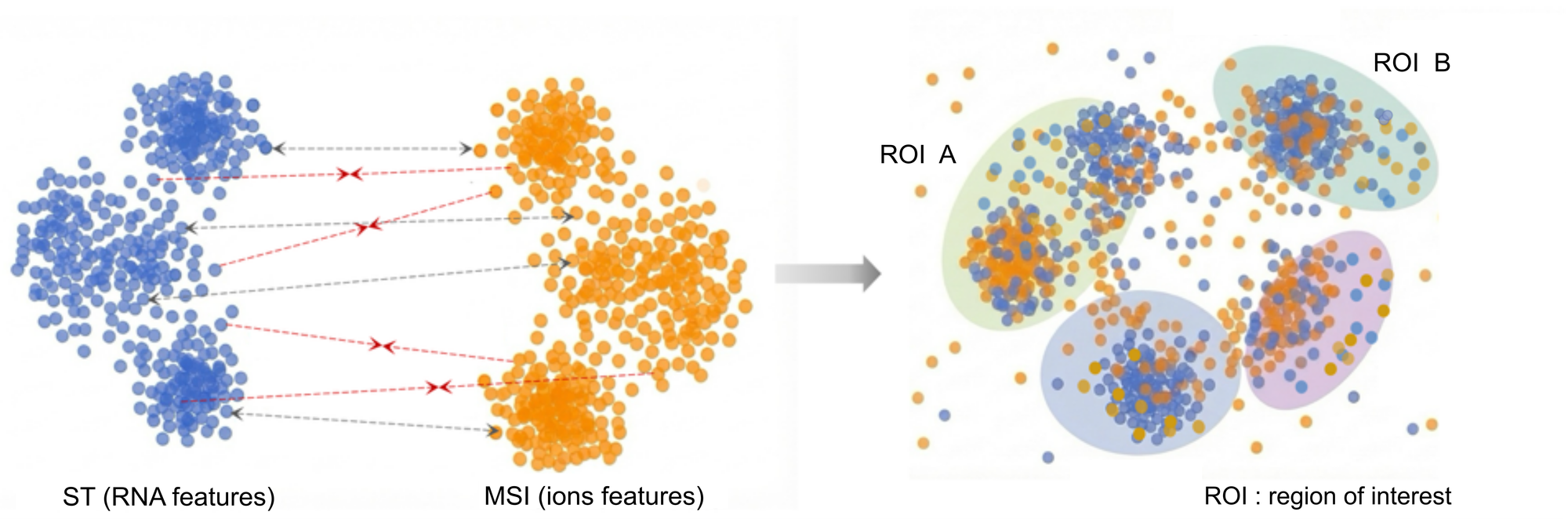
The objective of this internship is to develop a novel approach for semantic-based alignment of ST and MSI data, this is, a common representation space for joint embedding of two omics modalities. As a base for such ST+MSI common representation is an automatic supervised graph learning [5] of a shared embedding. This shared feature space can then be used for ST and SM joint embeddings, and subsequent ROI inference.
Candidate profile
The internship position is open to a Computer Science student: M2 or 3rd year of engineering school. Skills in ML are expected, and ideally, hands-on experience in deep-learning techniques applied to real data. Knowledge or experience on graph-learning is appreciated.
Excpected tasks
The intern will be responsible for:
- State-of-the-art Benchmarking: Evaluate existing multi-modal integration frameworks (e.g., SpatialGlue, GLUE, or Seurat v4) on public and in-house paired ST+MSI datasets to identify specific failure points in metabolic signal alignment.
- Methodological Development: Design and implement a novel Graph Neural Network (GNN) architecture to bridge the identified gaps.
- Implementation of Joint Embeddings: Develop an automatic supervised graph-learning workflow to map both modalities into a shared latent space, ensuring robustness against differences in spatial resolution and technical noise.
- Validation: Apply the developed model to in-house PDX glioblastoma datasets to characterize ROIs and validate the biological relevance of the integrated signal in collaboration with our cancer biology partners.
- Software Documentation: Deliver a clean, well-documented Python codebase (compatible with the lab’s existing DIMet/SpacePath ecosystem).
Supervision and location of the internship
The intern will be co-supervised by Dr Nikolski (Principal Investigator) and Johanna Galvis who recently defended her PhD. The internship position is for 6 months, at the Institut de Biochimie et Génétique Cellulaires (IBGC), under legal standard French gratification. The intern will have access to our HPC + GPU infrastructure. The multi-disciplinary nature of our team fosters knowledge sharing and skills upleveling.
How to apply
Interested candidates are invited to send a CV with a motivation letter to: macha.nikolski@u-bordeaux.fr and deisy-johanna.galvis-rodriguez@u-bordeaux.fr.
References
- Galvis J, Guyon J, Dartigues B, Hecht H, Grüning B, Specque F, Soueidan H, Karkar S, Daubon T, Nikolski M. DIMet: An Open-Source Tool for Differential Analysis of Targeted Isotope-Labeled Metabolomics Data. Bioinformatics 2024 40 (5) btae282. https://doi.org/10.1093/bioinformatics/btae282.
- Galvis J, Guyon J, Daubon T, Nikolski M. Using DIMet for Differential Analysis of Labeled Metabolomics Data: A Step-by-Step Guide Showcasing the Glioblastoma Metabolism. Bio-Protoc. 2025 15 (2) e5168. https://doi.org/10.21769/BioProtoc.5168.
- Ravi VM, Will P, Kueckelhaus J,… & Heiland DH Spatially Resolved Multi-Omics Deciphers Bidirectional Tumor-Host Interdependence in Glioblastoma. Cancer Cell 2022 40 (6) 639-655.e13. https://doi.org/10.1016/j.ccell.2022.05.009.
- Wheeler K, Gosmanov C, Jimenez Sandoval M, Yang Z, McCall LI. Frontiers in Mass Spectrometry-Based Spatial Metabolomics: Current Applications and Challenges in the Context of Biomedical Research. TrAC Trends Anal. Chem. 2024 175 117713. https://doi.org/10.1016/j.trac.2024.117713.
- Liu T, Fang Z.-Y, Zhang Z, Yu Y, Li M, Yin M.-Z. A Comprehensive Overview of Graph Neural Network-Based Approaches to Clustering for Spatial Transcriptomics. Comput. Struct. Biotechnol. J. 2024 23 106–128. https://doi.org/10.1016/j.csbj.2023.11.055.

