SeuratIntegrate: an R package to facilitate the use of integration methods with Seurat
Available on GitHub (cbib/Seurat-Integrate) and complemented by online documentation, examples, tutorials, and more. To bring the power of Scanorama, BBKNN, scVI and others to Seurat, give it a try!
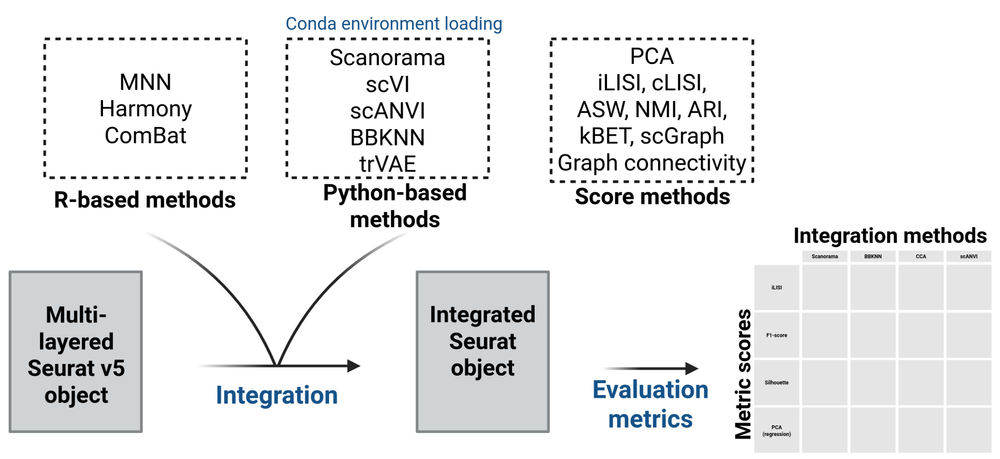
Single-cell RNA sequencing (scRNA-seq) has been in the spotlight for the last years - the number of publications have exponentially increased and the amount of publicly available cell transcriptomes has multiplied. Thus, integrating multiple datasets has become an increasingly common task in scRNA-seq analysis as exemplified by the advent of single-cell atlases that combine large amount of data from various sources. While Seurat offers access to batch-correction methods, the diversity of available options remains limited. With growing evidence that integration method performance varies significantly between datasets, making an informed decision in selecting the most appropriate integration approach is essential but not trivial. A broader range of accessible methods combined with a comprehensive toolbox for comparative integration analysis, would support more effective and flexible single-cell data integration workflows.
That is precisely what we assessed in our lab while integrating increasing number of scRNA-seq datasets. We felt that we missed options. Thus, we initially created wrappers for R integration methods to make them compatible with Seurat. Then, wrappers for Python methods. And finally, we designed functions to easily evaluate the performances of integrations. We embedded these functionalities in a R package we are thrilled to present: SeuratIntegrate. It will soon be published in Bioinformatics (Specque F. et al., 2025. doi: 10.1093/bioinformatics/btaf358).
Built on Seurat's foundations, SeuratIntegrate is an open source R package that expands integration methods available to Seurat users, including Python-based approaches, while operating entirely within the R environment. The package enables integration benchmarking using well-established performance metrics, and provides automated Python environment management, cross-language object conversion, and tools for score handling and visualization.
It is available on GitHub (cbib/Seurat-Integrate) and is complemented by a website with documentation, examples, tutorials, and more. To bring the power of Scanorama, BBKNN, scVI and others to Seurat, give it a try!


