DypFISH
Dynamic Patterned FISH to Interrogate RNA and Protein Spatial and Temporal Subcellular Distribution
Anca F. Savulescu, Robyn Brackin, Emmanuel Bouilhol, Benjamin Dartigues, Jonathan H. Warrell, Mafalda R. Pimentel, Nicolas Beaume, Isabela C. Fortunato, Stephane Dallongeville, Mikaël Boulle, Hayssam Soueidan, Fabrice Agou, Jan Schmoranzer, Jean-Christophe Olivo-Marin, Claudio A. Franco, Edgar R. Gomes, Macha Nikolski and Musa M. Mhlanga

Overview
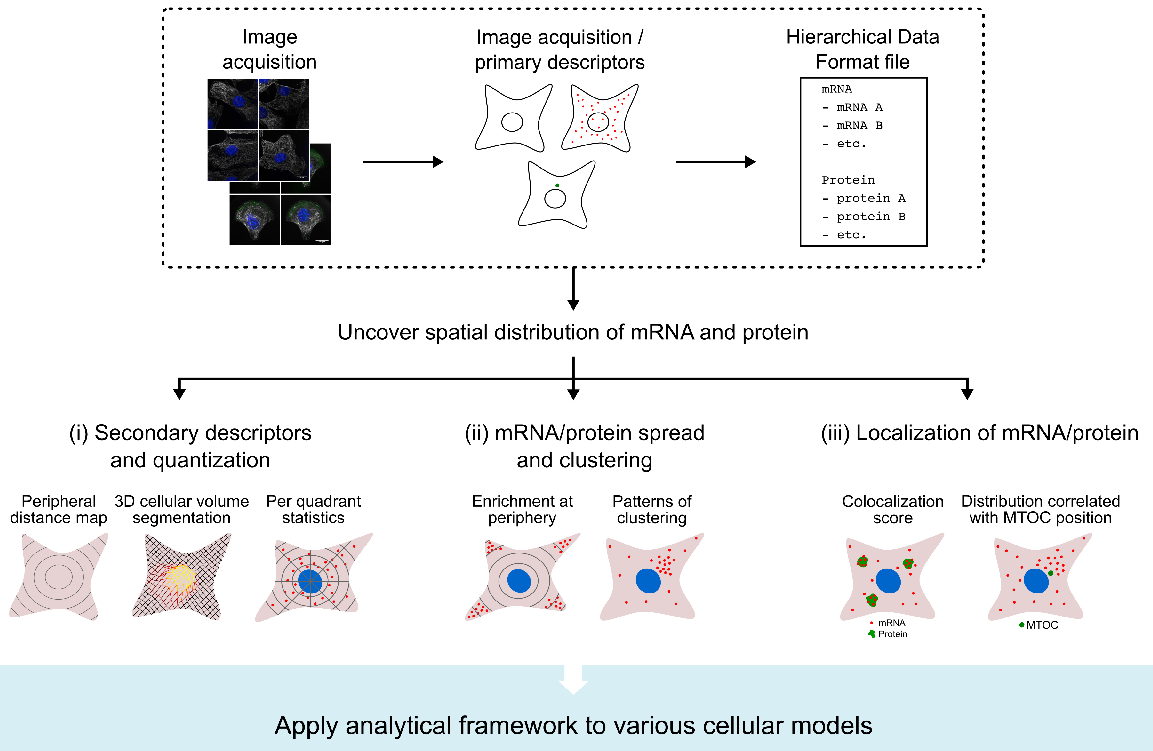
DypFISH is a spatially resolved high throughput approach for measuring quantitatively the spatial distribution of slow-scale dynamics of mRNA and protein distributions at fine-grained spatial resolution in single cells. DypFISH introduces analytical techniques that allow joint analysis of discrete point-based single molecule Fluorescence In Situ Hybridization (FISH) mRNA data and continuous intensity immunofluorescence (IF) protein data. The analytical techniques include a generalized approach to identifying clustering dynamics, an approach to identifying dependencies of mRNA and protein spatial distributions on organelle positioning and an approach to identifying interdependent dynamics between mRNA transcripts and their corresponding protein products globally and at specific subcellular locations.
Code
The source code of the downstream analysis pipeline can be downloaded from our GitHub
Data
| Dataset | Link |
|---|---|
| H5 files for Micropatterned Arhgdia, Gapdh, B-actin, Pard3, Rab13, Pkp4. Config file | Download |
| H5 file for CHX data, config file | Download |
| H5 file for CytoD data, config file | Download |
| H5 file for Nocodazole data, config file | Download |
| H5 file for PRRC2C data, config file | Download |
| H5 file for Muscle data, config file | Download |
Datasets to run the unit tests, as well as sample config : Download
Citation
DypFISH: Dynamic Patterned FISH to Interrogate RNA and Protein Spatial and Temporal Subcellular Distribution Anca F. Savulescu, Robyn Brackin, Emmanuel Bouilhol, Benjamin Dartigues, Jonathan H. Warrell, Mafalda R. Pimentel, Stephane Dallongeville, Jan Schmoranzer, Jean-Christophe Olivo-Marin, Edgar R. Gomes, Macha Nikolski, Musa M. Mhlanga bioRxiv 536383; doi: https://doi.org/10.1101/536383


